REFERENCES
- [1] R. Thompson, (2020) “Pandemic potential of 2019-nCoV" The Lancet Infectious Diseases 20(3): 280.
- [2] C. Chen, Y. DONG, J. KANG, S. LOU, F. WAN, H. LIU, and J. ZHANG, (2020) “Asymptomatic novel coronavirus pneumonia presenting as acute cerebral infarction: case report and review of the literature" Chinese Journal of Emergency Medicine: E017–E017.
- [3] V. S. Jagtap, P. More, and U. Jha, (2020) “A review of the 2019 novel coronavirus (COVID-19) based on current evidence":
- [4] Y. Fang, H. Zhang, J. Xie, M. Lin, L. Ying, P. Pang, andW. Ji, (2020) “Sensitivity of chest CT for COVID-19: comparison to RT-PCR" Radiology 296(2): E115–E117.
- [5] F. Khatami, M. Saatchi, S. S. T. Zadeh, Z. S. Aghamir, A. N. Shabestari, L. O. Reis, and S. M. K. Aghamir, (2020) “A meta-analysis of accuracy and sensitivity of chest CT and RT-PCR in COVID-19 diagnosis" Scientific reports 10(1): 1–12.
- [6] S. Yin and J. Bi. “Medical image annotation based on deep transfer learning”. In: 2018 IEEE International Conference on Internet of Things (iThings) and IEEE Green Computing and Communications (GreenCom) and IEEE Cyber, Physical and Social Computing (CPSCom) and IEEE Smart Data (SmartData). IEEE. 2018, 47–49.
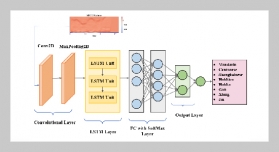
- [7] L. Teng, H. Li, and S. Karim, (2019) “DMCNN: A deep multiscale convolutional neural network model for medical image segmentation" Journal of Healthcare Engineering 2019:
- [8] A. D. Gritzman, M. Postema, D. M. Rubin, and V. Aharonson, (2021) “Threshold-based outer lip segmentation using support vector regression" Signal, Image and Video Processing: 1–6. DOI: 10.1007/s11760-020-01849-3.
- [9] M. Kowal, M. Z˙ ejmo, M. Skobel, J. Korbicz, and R. Monczak, (2020) “Cell nuclei segmentation in cytological images using convolutional neural network and seeded watershed algorithm" Journal of digital imaging 33(1): 231–242. DOI: 10.1007/s10278-019-00200-8.
- [10] S. Zhou and Z. Xu, (2020) “Automatic grayscale image segmentation based on affinity propagation clustering" Pattern Analysis And Applications 23(1): 331–348. DOI: 10.1007/s10044-019-00785-4.
- [11] S. Yin and H. Li, (2021) “GSAPSO-MQC: medical image encryption based on genetic simulated annealing particle swarm optimization and modified quantum chaos system" Evolutionary Intelligence 14(4): 1817–1829. DOI: 10.1007/s12065-020-00440-6.
- [12] S. Yin, H. Li, D. Liu, and S. Karim, (2020) “Active contour modal based on density-oriented BIRCH clustering method for medical image segmentation" Multimedia Tools and Applications 79(41): 31049–31068. DOI: 10.1007/s11042-020-09640-9.
- [13] J. Long, E. Shelhamer, and T. Darrell. “Fully convolutional networks for semantic segmentation”. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2015, 3431–3440. DOI: 10.1109/CVPR.2015.7298965.
- [14] O. Ronneberger, P. Fischer, and T. Brox. “U-net: Convolutional networks for biomedical image segmentation”. In: International Conference on Medical image computing and computer-assisted intervention. Springer. 2015, 234–241. DOI: 10.1007/978-3-319-24574-4_28.
- [15] F. Milletari, N. Navab, and S.-A. Ahmadi. “V-net: Fully convolutional neural networks for volumetric medical image segmentation”. In: 2016 fourth international conference on 3D vision (3DV). IEEE. 2016, 565–571. DOI: 10.1109/3DV.2016.79.
- [16] H. Zhao, J. Shi, X. Qi, X.Wang, and J. Jia. “Pyramid scene parsing network”. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2017, 6230–6239. DOI: 10.1109/CVPR.2017.660.
- [17] Y. Li and H. Chen. “Image recognition based on deep residual shrinkage Network”. In: 2021 International Conference on Artificial Intelligence and Electromechanical Automation (AIEA). IEEE. 2021, 334–337. DOI: 10.1109/AIEA53260.2021.00077..
- [18] B. QIAN, Z. XIAO, and W. SONG, (2020) “Application of Improved Convolutional Neural Network in Lung Image Segmentation" Journal of Frontiers of Computer Science and Technology 14(8): 1358–1367. DOI: 10.3778/.issn.1673-9418.2001042.
- [19] R. Zheng, Y. Zheng, and C. Dong-Ye, (2021) “Improved 3D U-Net for COVID-19 Chest CT Image Segmentation" Scientific Programming 2021: DOI: 10.1155/2021/9999368.
- [20] N. Gessert, M. Nielsen, M. Shaikh, R. Werner, and A. Schlaefer, (2020) “Skin lesion classification using ensembles of multi-resolution EfficientNets with meta data" MethodsX 7: 100864. DOI: 10 .1016/j.mex.2020.100864.
- [21] X.Wang, S. Yin, K. Sun, H. Li, J. Liu, S. Karim, et al., (2020) “GKFC-CNN: Modified Gaussian kernel fuzzy Cmeans and convolutional neural network for apple segmentation and recognition" Journal of Applied Science and Engineering 23(3): 555–561. DOI: 10.6180/jase.202009_23(3).0020.
- [22] L. Teng, H. Li, S. Yin, S. Karim, and Y. Sun, (2020) “An active contour model based on hybrid energy and fisher criterion for image segmentation" International Journal of Image and Data Fusion 11(1): 97–112. DOI: 10.1080/19479832.2019.1649309.
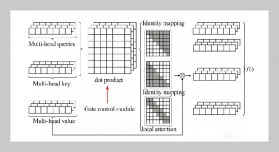
- [23] J. Hu, L. Shen, and G. Sun. “Squeeze-and-excitation networks”. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2018, 7132–7141. DOI: 10.1109/TPAMI.2019.2913372..
- [24] M. Li, Y.Wang, and C.Wang, (2021) “Recursive residual atrous spatial pyramid pooling network for single image deraining" Signal Processing: Image Communication 99: 116430. DOI: 10.1016/j.image.2021.116430.
- [25] URL: http://academictorrents.com/collection/lunalung-nodule-analysis16---isbi-2016-challenge/.
- [26] H. Chung, H. Ko, S. J. Jeon, K.-H. Yoon, and J. Lee, (2018) “Automatic lung segmentation with juxta-pleural nodule identification using active contour model and bayesian approach" IEEE journal of translational engineering in health and medicine 6: 1–13. DOI: 10.1109/JTEHM.2018.2837901.
- [27] L. Geng, S. Zhang, J. Tong, and Z. Xiao, (2019) “Lung segmentation method with dilated convolution based on VGG-16 network" Computer Assisted Surgery 24(sup2): 27–33. DOI: 10.1080/24699322.2019.1649071.
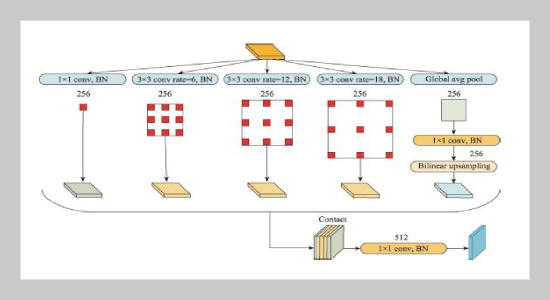
- [28] L.-C. Chen, G. Papandreou, F. Schroff, and H. Adam, (2017) “Rethinking atrous convolution for semantic image segmentation" arXiv preprint arXiv:1706.05587:
- [29] O. Oktay, J. Schlemper, L. L. Folgoc, M. Lee, M. Heinrich, K. Misawa, K. Mori, S. McDonagh, N. Y. Hammerla, B. Kainz, et al., (2018) “Attention u-net: Learning where to look for the pancreas" arXiv preprint arXiv:1804.03999:
- [30] A. Jaramillo-Yánez, M. E. Benalcázar, and E. Mena-Maldonado, (2020) “Real-time hand gesture recognition using surface electromyography and machine learning: A systematic literature review" Sensors 20(9): 2467. DOI: 10.3390/s20092467.
- [31] A. A. Khan, A. A. Laghari1, and S. A. Awan. Machine Learning in Computer Vision: A Review. EAI Endorsed Transactions on Scalable Information Systems. sis21(32):e4. DOI: http://dx.doi.org/10.4108/eai.21-4-2021.169418.