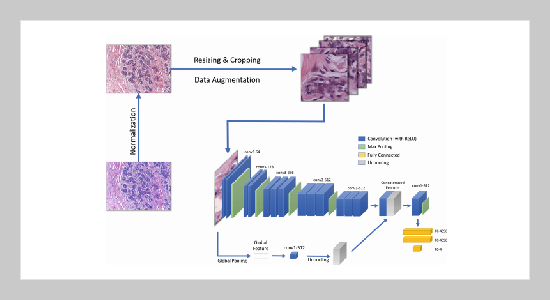
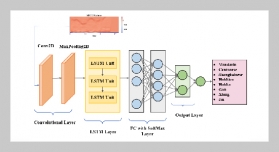
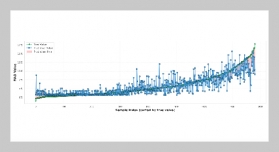
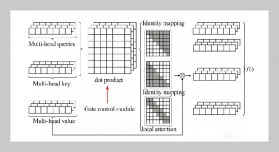
REFERENCES
- [1] J. Ferlay, M. Colombet, I. Soerjomataram, C. Mathers, D. M. Parkin, M. Piñeros, A. Znaor, and F. Bray. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. International Journal of Cancer, 144(8):1941–1953, 2019.
- [2] Alison Chetlen, Julie Mack, and Tiffany Chan. Breast cancer screening controversies: Who, when, why, and how? Clinical Imaging, 40(2):279–282, 2016.
- [3] Osamah Ibrahim Khalaf, Ghaida Muttashar Abdulsahib, and Bayan Mahdi Sabbar. Optimization of wireless sensor network coverage using the bee algorithm. Journal of Information Science and Engineering, 36(2):377– 386, 2020.
- [4] Andrei Chekkoury, Parmeshwar Khurd, Jie Ni, Claus Bahlmann, Ali Kamen, Amar Patel, Leo Grady, Maneesh Singh, Martin Groher, Nassir Navab, Elizabeth Krupinski, Jeffrey Johnson, Anna Graham, and Ronald Weinstein. Automated malignancy detection in breast histopathological images. Medical Imaging 2012: Computer-Aided Diagnosis, 8315:831515, 2012.
- [5] Ghaida Muttashar Abdulsahib and Osamah Ibrahim Khalaf. Comparison and evaluation of cloud processing models in cloud-based networks. International Journal of Simulation: Systems, Science and Technology, 19(5):26.1– 26.6, 2018.
- [6] Irene Pollanen, Billy Braithwaite, Tiia Ikonen, Harri Niska, Keijo Haataja, Pekka Toivanen, and Teemu Tolonen. Computer-aided breast cancer histopathological diagnosis: Comparative analysis of three DTOCSbased features: SW-DTOCS, SW-WDTOCS and SW-3-4- DTOCS. 2014 4th International Conference on Image Processing Theory, Tools and Applications, IPTA 2014, pages 0–5, 2015.
- [7] Pawel Filipczuk, Thomas Fevens, Adam Krzyzak, and Roman Monczak. Computer-aided breast cancer diagnosis based on the analysis of cytological images of fine needle biopsies. IEEE Transactions on Medical Imaging, 32(12):2169–2178, 2013.
- [8] T Mccann, John A. Ozolek, Carlos A. Castro, Michael T. McCann, John A. Ozolek, Carlos A. Castro, Bahram Parvin, and Jelena Kovaˇcevi´c. Automated Histology Analysis: Opportunities for signal processing. IEEE Signal Processing Magazine, (January):78–87, 2015.
- [9] World Health Organization (WHO). WHO Position paper on Mammography screening. World Health Organization, 2014.
- [10] Osamah Ibrahim Khalaf and Ghaida Muttashar Abdulsahib. Frequency estimation by the method of minimum mean squared error and P-value distributed in the wireless sensor network. Journal of Information Science and Engineering, 35(5):1099–1112, 2019.
- [11] Yoshua Bengio, Aaron Courville, and Pascal Vincent. Representation learning: A review and new perspectives. IEEE Transactions on Pattern Analysis and Machine Intelligence, 35(8):1798–1828, 2013.
- [12] Norrozila Sulaiman, Ghaidaa M. Abdulsahib, Osamah I. Khalaf, and Muamer N. Mohammed. Effect of using different propagations on performance of OLSR and DSDV routing protocols. In Proceedings - International Conference on Intelligent Systems, Modelling and Simulation, ISMS, volume 2015-Septe, pages 540–545, 2015.
- [13] Angel Alfonso Cruz-Roa, John Edison Arevalo Ovalle, Anant Madabhushi, and Fabio Augusto González Osorio. A deep learning architecture for image representation, visual interpretability and automated basal-cell carcinoma cancer detection. In Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), volume 8150 LNCS, pages 403–410, 2013.
- [14] Theyazn H.H. Aldhyani, Ali Saleh Alshebami, and Mohammed Y. Alzahrani. Soft Clustering for Enhancing the Diagnosis of Chronic Diseases over Machine Learning Algorithms. Journal of Healthcare Engineering, 2020, 2020.
- [15] Neeraj Dhungel, Gustavo Carneiro, and Andrew P. Bradley. Deep learning and structured prediction for the segmentation of mass in mammograms. In Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), volume 9349, pages 605–612. Springer Verlag, oct 2015.
- [16] Jeff Donahue, Yangqing Jia, Oriol Vinyals, Judy Hoffman, Ning Zhang, Eric Tzeng, and Trevor Darrell. DeCAF: A deep convolutional activation feature for generic visual recognition. In 31st International Conference on Machine Learning, ICML 2014, volume 2, pages 988–996, 2014.
- [17] Long D. Nguyen, Dongyun Lin, Zhiping Lin, and Jiuwen Cao. Deep CNNs for microscopic image classification by exploiting transfer learning and feature concatenation. In Proceedings - IEEE International Symposium on Circuits and Systems, volume 2018-May, 2018.
- [18] Ruqayya Awan, Navid Alemi Koohbanani, Muhammad Shaban, Anna Lisowska, and Nasir Rajpoot. Context-Aware Learning Using Transferable Features for Classification of Breast Cancer Histology Images. In Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), volume 10882 LNCS, pages 788–795. Springer Verlag, 2018.
- [19] Murat Karabatak. A new classifier for breast cancer detection based on Naïve Bayesian. Measurement: Journal of the International Measurement Confederation, 72:32–36, 2015.
- [20] Sana Ullah Khan, Naveed Islam, Zahoor Jan, Ikram Ud Din, and Joel J.P.C. Rodrigues. A novel deep learning based framework for the detection and classification of breast cancer using transfer learning. Pattern Recognition Letters, 125:1–6, 2019.
- [21] Fabio A. Spanhol, Paulo R. Cavalin, Luiz S. Oliveira, Caroline Petitjean, and Laurent Heutte. Deep features for breast cancer histopathological image classification. In 2017 IEEE International Conference on Systems, Man, and Cybernetics, SMC 2017, volume 2017-Janua, pages 1868–1873, 2017.
- [22] Qi Zhang, Yang Xiao, Wei Dai, Jingfeng Suo, Congzhi Wang, Jun Shi, and Hairong Zheng. Deep learning based classification of breast tumors with shear-wave elastography. Ultrasonics, 72:150–157, 2016.
- [23] Jun Xu, Lei Xiang, Qingshan Liu, Hannah Gilmore, Jianzhong Wu, Jinghai Tang, and Anant Madabhushi. Stacked sparse autoencoder (SSAE) for nuclei detection on breast cancer histopathology images. IEEE Transactions on Medical Imaging, 35(1):119–130, 2016.
- [24] Hiba Asri, Hajar Mousannif, Hassan Al Moatassime, and Thomas Noel. Using machine learning algorithms for breast cancer risk prediction and diagnosis. Procedia Computer Science, 83:1064–1069, 2016.
- [25] Arpit Bhardwaj and Aruna Tiwari. Breast cancer diagnosis using Genetically Optimized Neural Network model. Expert Systems with Applications, 42(10):4611– 4620, 2015.
- [26] Yao Guo, Huihui Dong, Fangzhou Song, Chuang Zhu, and Jun Liu. Breast Cancer Histology Image Classification Based on Deep Neural Networks. In Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), volume 10882 LNCS, pages 827–836. Springer Verlag, 2018.
- [27] Sulaiman Vesal, Nishant Ravikumar, Amir Abbas Davari, Stephan Ellmann, and Andreas Maier. Classification of Breast Cancer Histology Images Using Transfer Learning. In Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), volume 10882 LNCS, pages 812–819. Springer Verlag, 2018.
- [28] Alex Krizhevsky, Ilya Sutskever, and Geoffrey E Hinton. ImageNet classification with deep convolutional neural networks. Communications of the ACM, 60(6):84– 90, jun 2017.
- [29] Dan C. Cire¸san, Alessandro Giusti, Luca M. Gambardella, and Jürgen Schmidhuber. Mitosis detection in breast cancer histology images with deep neural networks. In Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), volume 8150 LNCS, pages 411–418, 2013.
- [30] Marc Macenko, Marc Niethammer, J. S. Marron, David Borland, John T. Woosley, Xiaojun Guan, Charles Schmitt, and Nancy E. Thomas. A method for normalizing histology slides for quantitative analysis. In Proceedings - 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, ISBI 2009, pages 1107– 1110, 2009.
- [31] Kosmas Dimitropoulos, Panagiotis Barmpoutis, Christina Zioga, Athanasios Kamas, Kalliopi Patsiaoura, and Nikos Grammalidis. Grading of invasive breast carcinoma through Grassmannian VLAD encoding. PLoS ONE, 12(9), sep 2017.
- [32] Fabio A. Spanhol, Luiz S. Oliveira, Caroline Petitjean, and Laurent Heutte. A Dataset for Breast Cancer Histopathological Image Classification. IEEE Transactions on Biomedical Engineering, 63(7):1455–1462, 2016.
- [33] Duc My Vo, Ngoc Quang Nguyen, and Sang Woong Lee. Classification of breast cancer histology images using incremental boosting convolution networks. Information Sciences, 482:123–138, 2019.
- [34] Fabio A Spanhol, Luiz S Oliveira, Caroline Petitjean, and Laurent Heutte. A dataset for breast cancer histopathological image classification. IEEE Transactions on Biomedical Engineering, 63(7):1455–1462, 2015.
- [35] David West and Vivian West. Model selection for a medical diagnostic decision support system: A breast cancer detection case. Artificial Intelligence in Medicine, 20(3):183–204, 2000.
- [36] Neslihan Bayramoglu, Juho Kannala, and Janne Heikkila. Deep learning for magnification independent breast cancer histopathology image classification. In Proceedings - International Conference on Pattern Recognition, pages 2440–2445, 2016.
- [37] Mohamad Mahmoud Al Rahhal. Breast cancer classification in histopathological images using convolutional neural network. International Journal of Advanced Computer Science and Applications, 9(3):64–68, 2018.